
Calculate fragment euclidean nearest neighbor distance
Source:R/lsm_enn_distance.R, R/lsm_enn_distance_vector.R
lsm_enn_distance.RdCalculate euclidean nearest neighbor distance among fragments in meters using r.clump and r.distance GRASS GIS module.
Calculate euclidean nearest neighbor distance among fragments in meters using r.distance GRASS GIS module.
Usage
lsm_enn_distance(input, output = NULL, directions = 8, zero_as_na = FALSE)
lsm_enn_distance(input, output = NULL, directions = 8, zero_as_na = FALSE)Arguments
- input
[character=""]
Habitat map, following a binary classification (e.g. values 1,0 or 1,NA for habitat,non-habitat).- output
[character=""]
fragment fragment euclidean nearest neighbor distance map name inside GRASS Data Base.- directions
[numerical=""]- zero_as_na
[logical=""]- export_table
[logical=""]
Examples
library(lsmetrics)
library(terra)
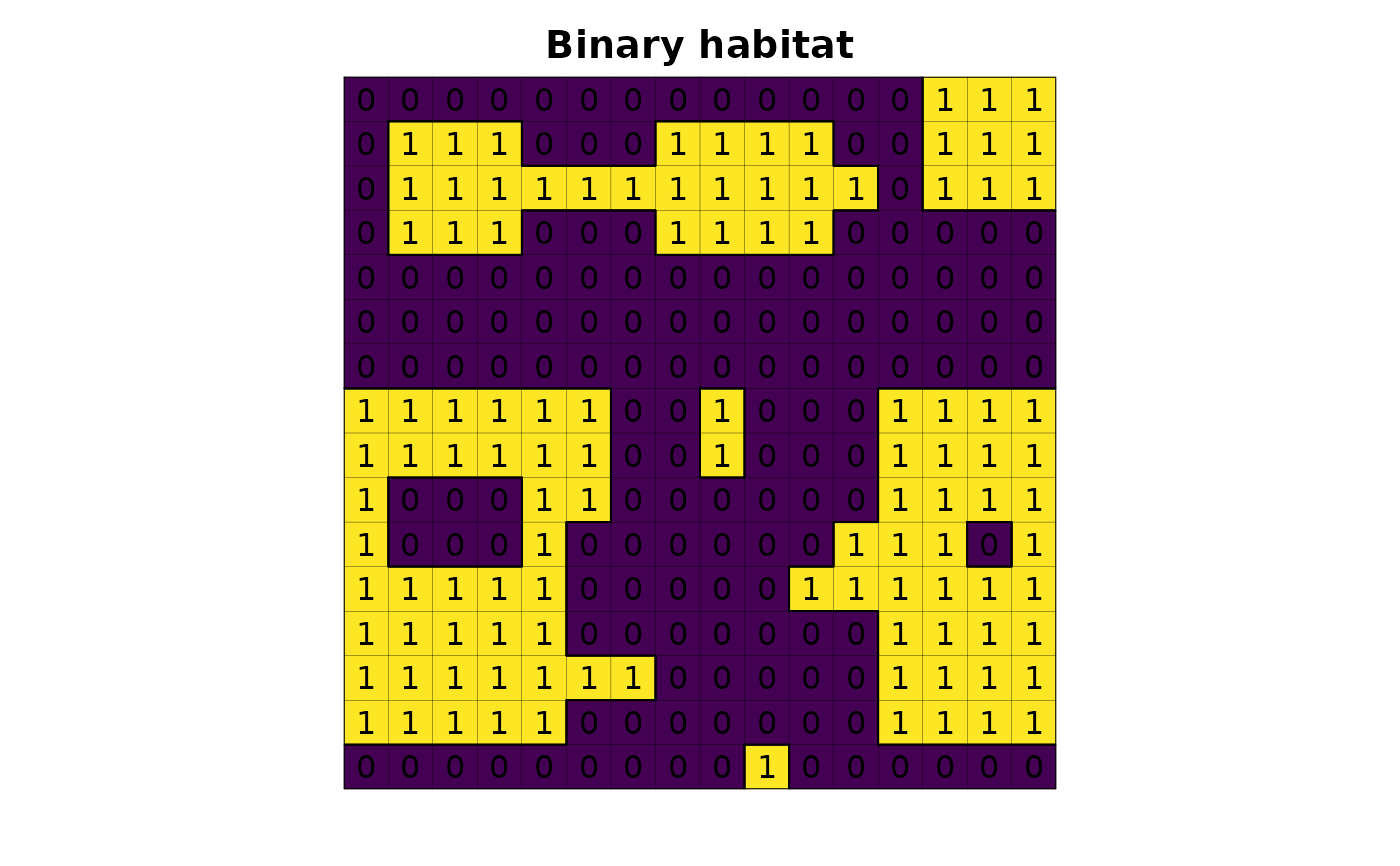
# read habitat data
f <- system.file("raster/toy_landscape_habitat.tif", package = "lsmetrics")
r <- terra::rast(f)
# plot
plot(r, legend = FALSE, axes = FALSE, main = "Binary habitat")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r)
 # find grass
path_grass <- system("grass --config path", inter = TRUE) # windows users need to find the grass gis path installation, e.g. "C:/Program Files/GRASS GIS 8.3"
# create grassdb
rgrass::initGRASS(gisBase = path_grass,
SG = r,
gisDbase = "grassdb",
location = "newLocation",
mapset = "PERMANENT",
override = TRUE)
#> gisdbase grassdb
#> location newLocation
#> mapset PERMANENT
#> rows 16
#> columns 16
#> north 7525600
#> south 7524000
#> west 234000
#> east 235600
#> nsres 100
#> ewres 100
#> projection:
#> PROJCRS["WGS 84 / UTM zone 23S",
#> BASEGEOGCRS["WGS 84",
#> ENSEMBLE["World Geodetic System 1984 ensemble",
#> MEMBER["World Geodetic System 1984 (Transit)"],
#> MEMBER["World Geodetic System 1984 (G730)"],
#> MEMBER["World Geodetic System 1984 (G873)"],
#> MEMBER["World Geodetic System 1984 (G1150)"],
#> MEMBER["World Geodetic System 1984 (G1674)"],
#> MEMBER["World Geodetic System 1984 (G1762)"],
#> MEMBER["World Geodetic System 1984 (G2139)"],
#> ELLIPSOID["WGS 84",6378137,298.257223563,
#> LENGTHUNIT["metre",1]],
#> ENSEMBLEACCURACY[2.0]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433]],
#> ID["EPSG",4326]],
#> CONVERSION["UTM zone 23S",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",0,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-45,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",500000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",10000000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["(E)",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1]],
#> AXIS["(N)",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1]],
#> USAGE[
#> SCOPE["Navigation and medium accuracy spatial referencing."],
#> AREA["Between 48°W and 42°W, southern hemisphere between 80°S and equator, onshore and offshore. Brazil."],
#> BBOX[-80,-48,0,-42]],
#> ID["EPSG",32723]]
# import raster from r to grass
rgrass::write_RAST(x = r, flags = c("o", "overwrite", "quiet"), vname = "r")
#> SpatRaster read into GRASS using r.in.gdal from file
# distance
lsmetrics::lsm_enn_distance(input = "r", zero_as_na = FALSE)
#> Converting zero as null
#> Identifying fragments
#> Calculating distance
#> Changing the raster color
#> Cleaning files
#> Warning: The command:
#> g.remove -b -f --quiet type=raster name=r_enn_distance_null,r_enn_distance_clump,r_enn_distance_temp
#> produced at least one warning during execution:
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
# files
rgrass::execGRASS(cmd = "g.list", type = "raster")
#> r
#> r_enn_distance
# import from grass to r
r_enn_distance <- terra::rast(rgrass::read_RAST("r_enn_distance", flags = "quiet", return_format = "SGDF"))
#> Creating BIL support files...
#> Exporting raster as integer values (bytes=4)
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
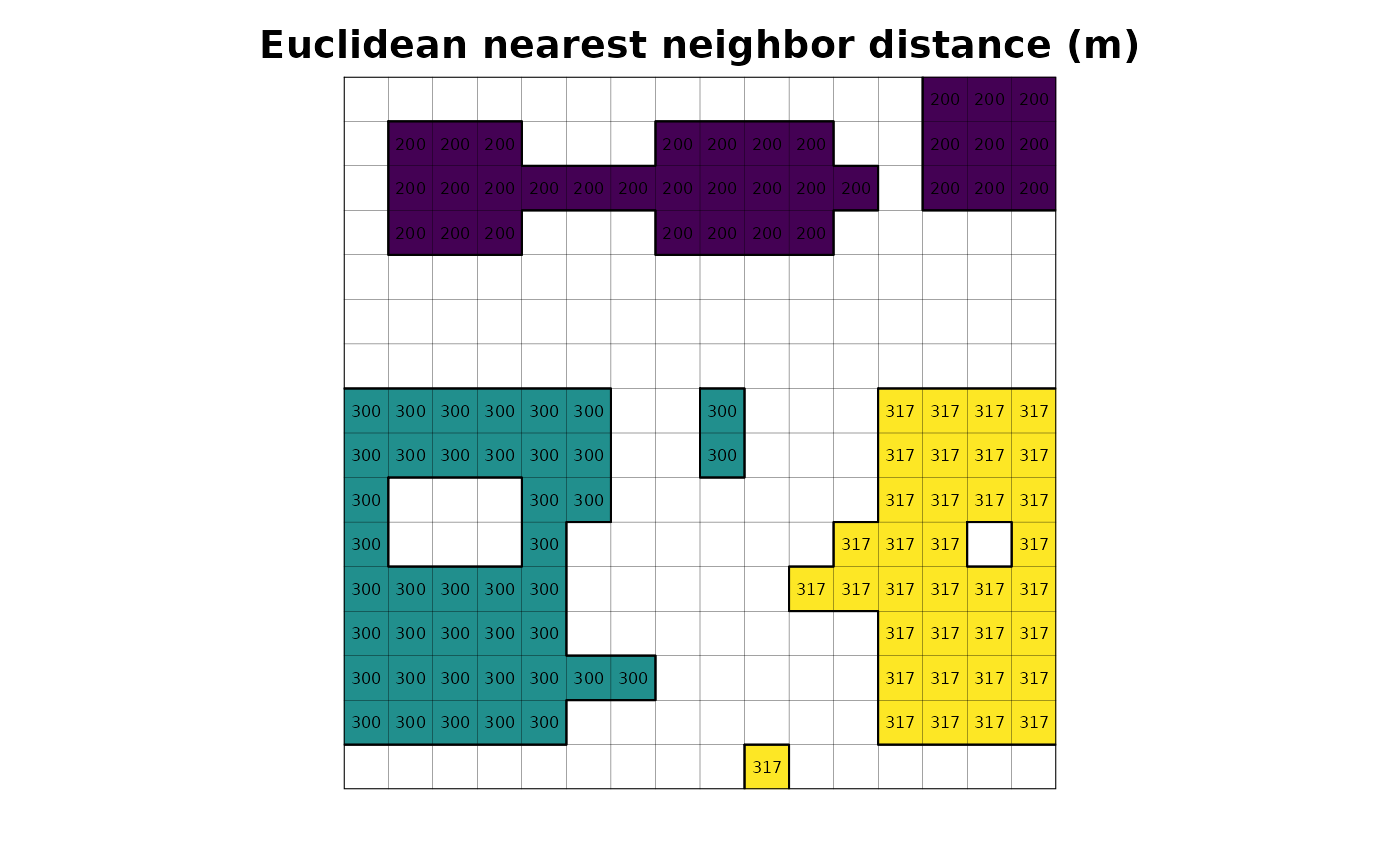
# plot
plot(r_enn_distance, legend = FALSE, axes = FALSE, main = "Euclidean nearest neighbor distance (m)")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r_enn_distance, cex = .5)
# find grass
path_grass <- system("grass --config path", inter = TRUE) # windows users need to find the grass gis path installation, e.g. "C:/Program Files/GRASS GIS 8.3"
# create grassdb
rgrass::initGRASS(gisBase = path_grass,
SG = r,
gisDbase = "grassdb",
location = "newLocation",
mapset = "PERMANENT",
override = TRUE)
#> gisdbase grassdb
#> location newLocation
#> mapset PERMANENT
#> rows 16
#> columns 16
#> north 7525600
#> south 7524000
#> west 234000
#> east 235600
#> nsres 100
#> ewres 100
#> projection:
#> PROJCRS["WGS 84 / UTM zone 23S",
#> BASEGEOGCRS["WGS 84",
#> ENSEMBLE["World Geodetic System 1984 ensemble",
#> MEMBER["World Geodetic System 1984 (Transit)"],
#> MEMBER["World Geodetic System 1984 (G730)"],
#> MEMBER["World Geodetic System 1984 (G873)"],
#> MEMBER["World Geodetic System 1984 (G1150)"],
#> MEMBER["World Geodetic System 1984 (G1674)"],
#> MEMBER["World Geodetic System 1984 (G1762)"],
#> MEMBER["World Geodetic System 1984 (G2139)"],
#> ELLIPSOID["WGS 84",6378137,298.257223563,
#> LENGTHUNIT["metre",1]],
#> ENSEMBLEACCURACY[2.0]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433]],
#> ID["EPSG",4326]],
#> CONVERSION["UTM zone 23S",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",0,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-45,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",500000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",10000000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["(E)",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1]],
#> AXIS["(N)",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1]],
#> USAGE[
#> SCOPE["Navigation and medium accuracy spatial referencing."],
#> AREA["Between 48°W and 42°W, southern hemisphere between 80°S and equator, onshore and offshore. Brazil."],
#> BBOX[-80,-48,0,-42]],
#> ID["EPSG",32723]]
# import raster from r to grass
rgrass::write_RAST(x = r, flags = c("o", "overwrite", "quiet"), vname = "r")
#> SpatRaster read into GRASS using r.in.gdal from file
# distance
lsmetrics::lsm_enn_distance(input = "r", zero_as_na = FALSE)
#> Converting zero as null
#> Identifying fragments
#> Calculating distance
#> Changing the raster color
#> Cleaning files
#> Warning: The command:
#> g.remove -b -f --quiet type=raster name=r_enn_distance_null,r_enn_distance_clump,r_enn_distance_temp
#> produced at least one warning during execution:
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
# files
rgrass::execGRASS(cmd = "g.list", type = "raster")
#> r
#> r_enn_distance
# import from grass to r
r_enn_distance <- terra::rast(rgrass::read_RAST("r_enn_distance", flags = "quiet", return_format = "SGDF"))
#> Creating BIL support files...
#> Exporting raster as integer values (bytes=4)
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
# plot
plot(r_enn_distance, legend = FALSE, axes = FALSE, main = "Euclidean nearest neighbor distance (m)")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r_enn_distance, cex = .5)
 # delete grassdb
unlink("grassdb", recursive = TRUE)
library(lsmetrics)
library(terra)
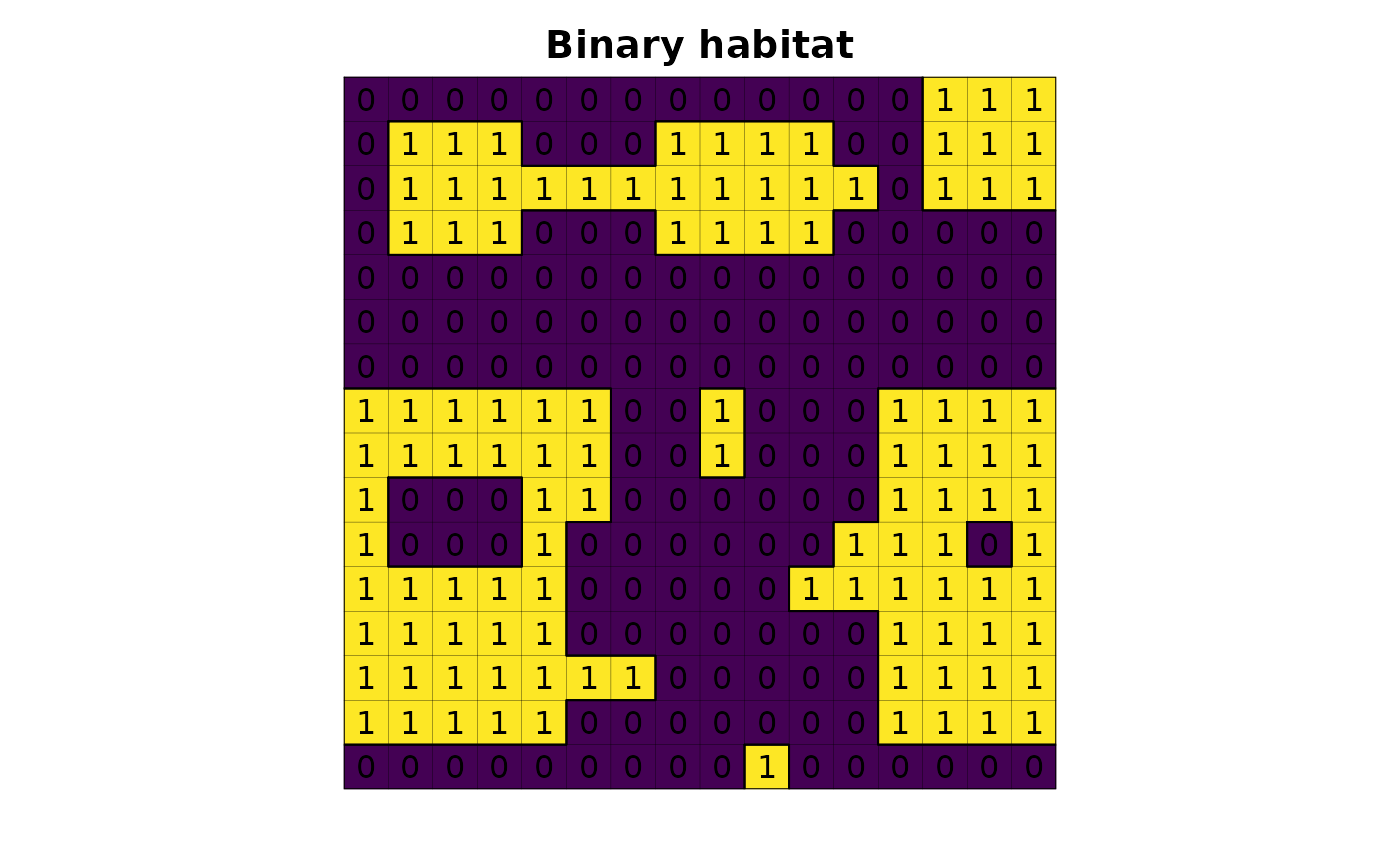
# read habitat data
f <- system.file("raster/toy_landscape_habitat.tif", package = "lsmetrics")
r <- terra::rast(f)
# plot
plot(r, legend = FALSE, axes = FALSE, main = "Binary habitat")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r)
# delete grassdb
unlink("grassdb", recursive = TRUE)
library(lsmetrics)
library(terra)
# read habitat data
f <- system.file("raster/toy_landscape_habitat.tif", package = "lsmetrics")
r <- terra::rast(f)
# plot
plot(r, legend = FALSE, axes = FALSE, main = "Binary habitat")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r)
 # find grass
path_grass <- system("grass --config path", inter = TRUE) # windows users need to find the grass gis path installation, e.g. "C:/Program Files/GRASS GIS 8.3"
# create grassdb
rgrass::initGRASS(gisBase = path_grass,
SG = r,
gisDbase = "grassdb",
location = "newLocation",
mapset = "PERMANENT",
override = TRUE)
#> gisdbase grassdb
#> location newLocation
#> mapset PERMANENT
#> rows 16
#> columns 16
#> north 7525600
#> south 7524000
#> west 234000
#> east 235600
#> nsres 100
#> ewres 100
#> projection:
#> PROJCRS["WGS 84 / UTM zone 23S",
#> BASEGEOGCRS["WGS 84",
#> ENSEMBLE["World Geodetic System 1984 ensemble",
#> MEMBER["World Geodetic System 1984 (Transit)"],
#> MEMBER["World Geodetic System 1984 (G730)"],
#> MEMBER["World Geodetic System 1984 (G873)"],
#> MEMBER["World Geodetic System 1984 (G1150)"],
#> MEMBER["World Geodetic System 1984 (G1674)"],
#> MEMBER["World Geodetic System 1984 (G1762)"],
#> MEMBER["World Geodetic System 1984 (G2139)"],
#> ELLIPSOID["WGS 84",6378137,298.257223563,
#> LENGTHUNIT["metre",1]],
#> ENSEMBLEACCURACY[2.0]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433]],
#> ID["EPSG",4326]],
#> CONVERSION["UTM zone 23S",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",0,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-45,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",500000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",10000000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["(E)",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1]],
#> AXIS["(N)",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1]],
#> USAGE[
#> SCOPE["Navigation and medium accuracy spatial referencing."],
#> AREA["Between 48°W and 42°W, southern hemisphere between 80°S and equator, onshore and offshore. Brazil."],
#> BBOX[-80,-48,0,-42]],
#> ID["EPSG",32723]]
# import raster from r to grass
rgrass::write_RAST(x = r, flags = c("o", "overwrite", "quiet"), vname = "r")
#> SpatRaster read into GRASS using r.in.gdal from file
# distance
lsmetrics::lsm_enn_distance(input = "r", zero_as_na = FALSE)
#> Converting zero as null
#> Identifying fragments
#> Calculating distance
#> Changing the raster color
#> Cleaning files
#> Warning: The command:
#> g.remove -b -f --quiet type=raster name=r_enn_distance_null,r_enn_distance_clump,r_enn_distance_temp
#> produced at least one warning during execution:
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
# files
rgrass::execGRASS(cmd = "g.list", type = "raster")
#> r
#> r_enn_distance
# import from grass to r
r_enn_distance <- terra::rast(rgrass::read_RAST("r_enn_distance", flags = "quiet", return_format = "SGDF"))
#> Creating BIL support files...
#> Exporting raster as integer values (bytes=4)
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
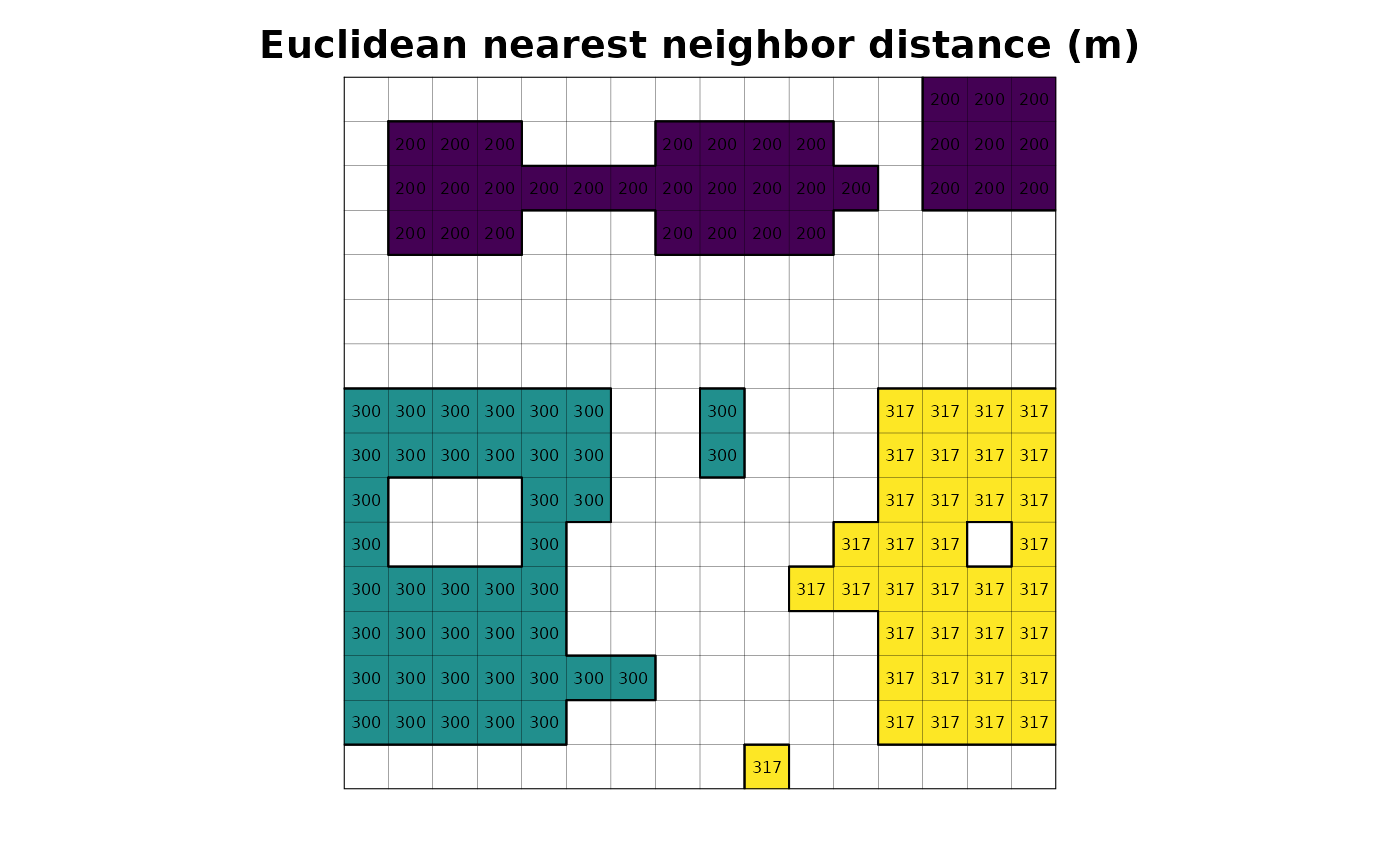
# plot
plot(r_enn_distance, legend = FALSE, axes = FALSE, main = "Euclidean nearest neighbor distance (m)")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r_enn_distance, cex = .5)
# find grass
path_grass <- system("grass --config path", inter = TRUE) # windows users need to find the grass gis path installation, e.g. "C:/Program Files/GRASS GIS 8.3"
# create grassdb
rgrass::initGRASS(gisBase = path_grass,
SG = r,
gisDbase = "grassdb",
location = "newLocation",
mapset = "PERMANENT",
override = TRUE)
#> gisdbase grassdb
#> location newLocation
#> mapset PERMANENT
#> rows 16
#> columns 16
#> north 7525600
#> south 7524000
#> west 234000
#> east 235600
#> nsres 100
#> ewres 100
#> projection:
#> PROJCRS["WGS 84 / UTM zone 23S",
#> BASEGEOGCRS["WGS 84",
#> ENSEMBLE["World Geodetic System 1984 ensemble",
#> MEMBER["World Geodetic System 1984 (Transit)"],
#> MEMBER["World Geodetic System 1984 (G730)"],
#> MEMBER["World Geodetic System 1984 (G873)"],
#> MEMBER["World Geodetic System 1984 (G1150)"],
#> MEMBER["World Geodetic System 1984 (G1674)"],
#> MEMBER["World Geodetic System 1984 (G1762)"],
#> MEMBER["World Geodetic System 1984 (G2139)"],
#> ELLIPSOID["WGS 84",6378137,298.257223563,
#> LENGTHUNIT["metre",1]],
#> ENSEMBLEACCURACY[2.0]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433]],
#> ID["EPSG",4326]],
#> CONVERSION["UTM zone 23S",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",0,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-45,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",500000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",10000000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["(E)",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1]],
#> AXIS["(N)",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1]],
#> USAGE[
#> SCOPE["Navigation and medium accuracy spatial referencing."],
#> AREA["Between 48°W and 42°W, southern hemisphere between 80°S and equator, onshore and offshore. Brazil."],
#> BBOX[-80,-48,0,-42]],
#> ID["EPSG",32723]]
# import raster from r to grass
rgrass::write_RAST(x = r, flags = c("o", "overwrite", "quiet"), vname = "r")
#> SpatRaster read into GRASS using r.in.gdal from file
# distance
lsmetrics::lsm_enn_distance(input = "r", zero_as_na = FALSE)
#> Converting zero as null
#> Identifying fragments
#> Calculating distance
#> Changing the raster color
#> Cleaning files
#> Warning: The command:
#> g.remove -b -f --quiet type=raster name=r_enn_distance_null,r_enn_distance_clump,r_enn_distance_temp
#> produced at least one warning during execution:
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
#> WARNING: Raster map <r_enn_distance_clump@PERMANENT> is a base map for
#> <r_enn_distance_temp@PERMANENT>. Remove forced.
# files
rgrass::execGRASS(cmd = "g.list", type = "raster")
#> r
#> r_enn_distance
# import from grass to r
r_enn_distance <- terra::rast(rgrass::read_RAST("r_enn_distance", flags = "quiet", return_format = "SGDF"))
#> Creating BIL support files...
#> Exporting raster as integer values (bytes=4)
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
# plot
plot(r_enn_distance, legend = FALSE, axes = FALSE, main = "Euclidean nearest neighbor distance (m)")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r_enn_distance, cex = .5)
 # delete grassdb
unlink("grassdb", recursive = TRUE)
# delete grassdb
unlink("grassdb", recursive = TRUE)