
Calculate fragment euclidean nearest neighbor distance
Source:R/lsm_distance_enn.R
lsm_distance_enn.RdCalculate euclidean nearest neighbor distance among fragments in meters using r.clump and r.distance GRASS GIS module.
Usage
lsm_distance_enn(
input,
output = NULL,
zero_as_null = FALSE,
region_input = FALSE,
id_direction = 8,
distance_round_digit = 0,
grid_size = 10000,
distance_radius = 2000,
table_distance = FALSE
)Arguments
- input
[character=""]
Habitat map, following a binary classification (e.g. values 1,0 or 1,NA for habitat,non-habitat).- output
[character=""]
Fragment fragment euclidean nearest neighbor distance map name inside GRASS Data Base.- zero_as_null
[logical=""]- table_distance
[logical=""]- id_directions
[numerical=""]
Examples
library(lsmetrics)
library(terra)
# read habitat data
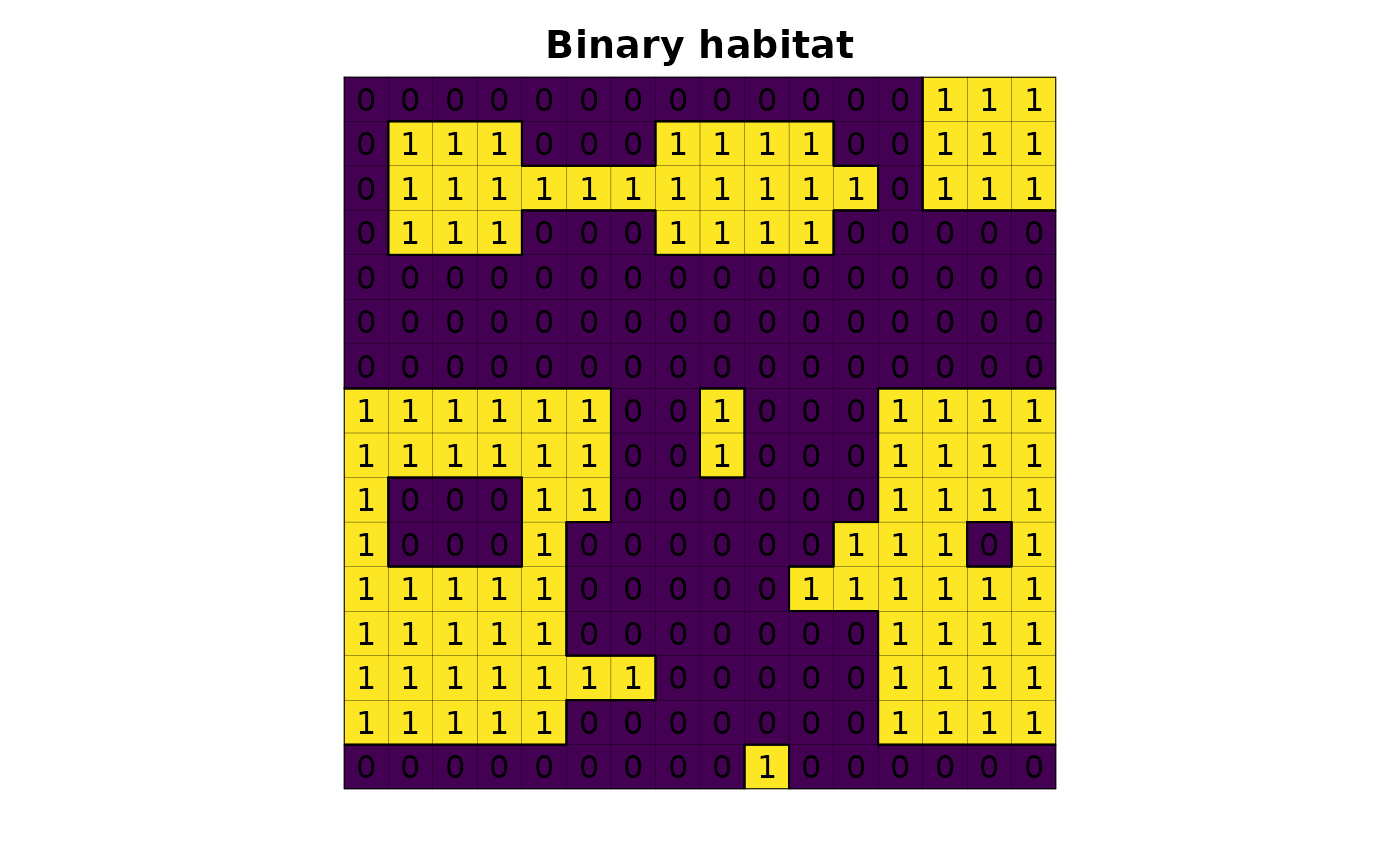
r <- lsmetrics::lsm_toy_landscape(proj_type = "meters")
# plot
plot(r, legend = FALSE, axes = FALSE, main = "Binary habitat")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r)
 # find grass
path_grass <- system("grass --config path", inter = TRUE) # windows users need to find the grass gis path installation, e.g. "C:/Program Files/GRASS GIS 8.3"
# create grassdb
rgrass::initGRASS(gisBase = path_grass,
SG = r,
gisDbase = "grassdb",
location = "newLocation",
mapset = "PERMANENT",
override = TRUE)
#> gisdbase grassdb
#> location newLocation
#> mapset PERMANENT
#> rows 16
#> columns 16
#> north 7525600
#> south 7524000
#> west 234000
#> east 235600
#> nsres 100
#> ewres 100
#> projection:
#> PROJCRS["WGS 84 / UTM zone 23S",
#> BASEGEOGCRS["WGS 84",
#> ENSEMBLE["World Geodetic System 1984 ensemble",
#> MEMBER["World Geodetic System 1984 (Transit)"],
#> MEMBER["World Geodetic System 1984 (G730)"],
#> MEMBER["World Geodetic System 1984 (G873)"],
#> MEMBER["World Geodetic System 1984 (G1150)"],
#> MEMBER["World Geodetic System 1984 (G1674)"],
#> MEMBER["World Geodetic System 1984 (G1762)"],
#> MEMBER["World Geodetic System 1984 (G2139)"],
#> ELLIPSOID["WGS 84",6378137,298.257223563,
#> LENGTHUNIT["metre",1]],
#> ENSEMBLEACCURACY[2.0]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433]],
#> ID["EPSG",4326]],
#> CONVERSION["UTM zone 23S",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",0,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-45,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",500000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",10000000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["(E)",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1]],
#> AXIS["(N)",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1]],
#> USAGE[
#> SCOPE["Navigation and medium accuracy spatial referencing."],
#> AREA["Between 48°W and 42°W, southern hemisphere between 80°S and equator, onshore and offshore. Brazil."],
#> BBOX[-80,-48,0,-42]],
#> ID["EPSG",32723]]
# import raster from r to grass
rgrass::write_RAST(x = r, flags = c("o", "overwrite", "quiet"), vname = "r")
#> SpatRaster read into GRASS using r.in.gdal from memory
# distance
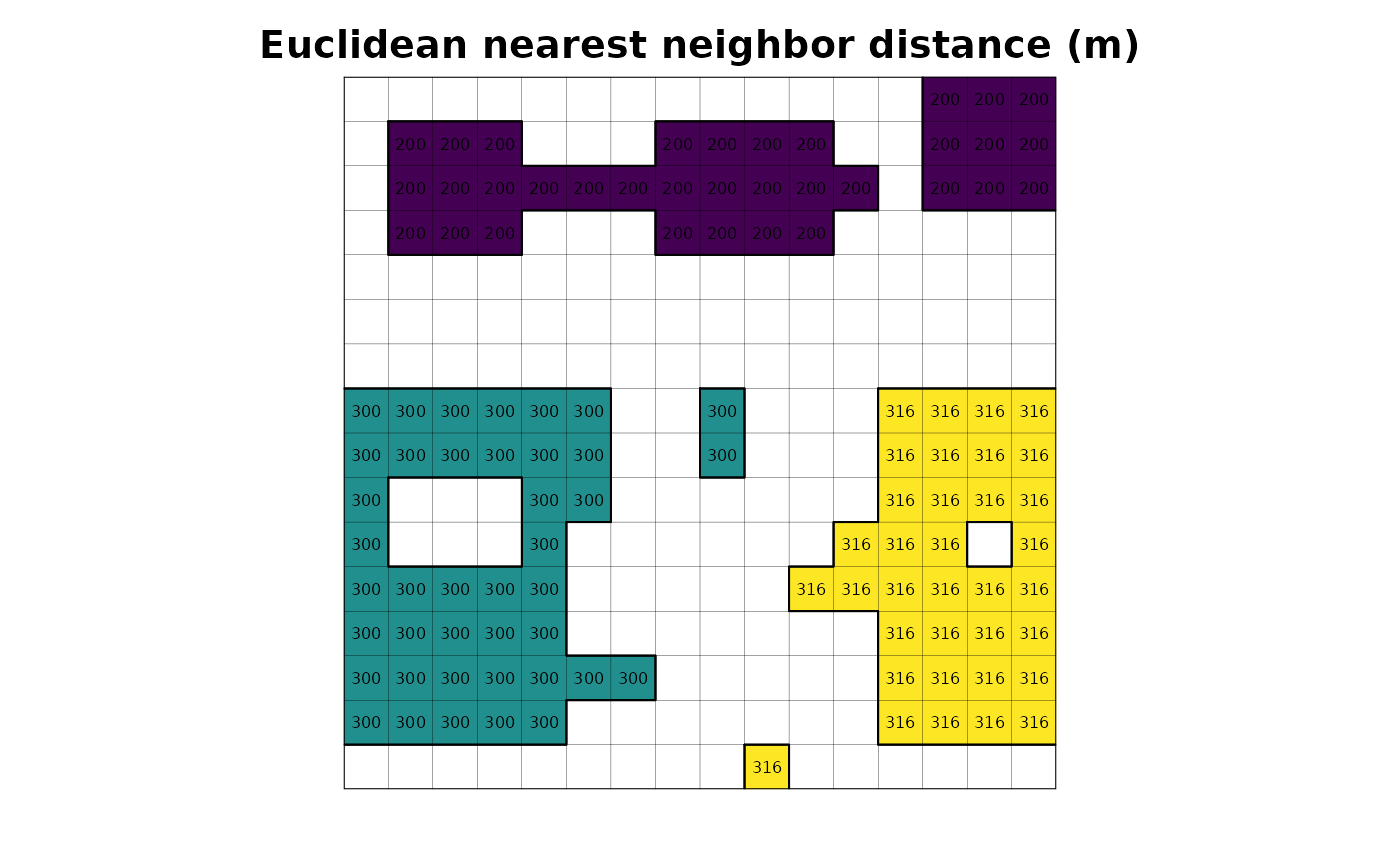
lsmetrics::lsm_distance_enn(input = "r")
#> Converting zero as null
#> Identifying fragments
#> Calculating distance
#> [1] "1 of 1"
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
#> Changing the raster color
#> Cleaning data
# files
rgrass::execGRASS(cmd = "g.list", type = "raster")
#> r
#> r_distance_enn
# import from grass to r
r_distance_enn <- terra::rast(rgrass::read_RAST("r_distance_enn", flags = "quiet", return_format = "SGDF"))
#> Creating BIL support files...
#> Exporting raster as integer values (bytes=4)
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
# plot
plot(r_distance_enn, legend = FALSE, axes = FALSE, main = "Euclidean nearest neighbor distance (m)")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r_distance_enn, cex = .5)
# find grass
path_grass <- system("grass --config path", inter = TRUE) # windows users need to find the grass gis path installation, e.g. "C:/Program Files/GRASS GIS 8.3"
# create grassdb
rgrass::initGRASS(gisBase = path_grass,
SG = r,
gisDbase = "grassdb",
location = "newLocation",
mapset = "PERMANENT",
override = TRUE)
#> gisdbase grassdb
#> location newLocation
#> mapset PERMANENT
#> rows 16
#> columns 16
#> north 7525600
#> south 7524000
#> west 234000
#> east 235600
#> nsres 100
#> ewres 100
#> projection:
#> PROJCRS["WGS 84 / UTM zone 23S",
#> BASEGEOGCRS["WGS 84",
#> ENSEMBLE["World Geodetic System 1984 ensemble",
#> MEMBER["World Geodetic System 1984 (Transit)"],
#> MEMBER["World Geodetic System 1984 (G730)"],
#> MEMBER["World Geodetic System 1984 (G873)"],
#> MEMBER["World Geodetic System 1984 (G1150)"],
#> MEMBER["World Geodetic System 1984 (G1674)"],
#> MEMBER["World Geodetic System 1984 (G1762)"],
#> MEMBER["World Geodetic System 1984 (G2139)"],
#> ELLIPSOID["WGS 84",6378137,298.257223563,
#> LENGTHUNIT["metre",1]],
#> ENSEMBLEACCURACY[2.0]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433]],
#> ID["EPSG",4326]],
#> CONVERSION["UTM zone 23S",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",0,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-45,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",500000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",10000000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["(E)",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1]],
#> AXIS["(N)",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1]],
#> USAGE[
#> SCOPE["Navigation and medium accuracy spatial referencing."],
#> AREA["Between 48°W and 42°W, southern hemisphere between 80°S and equator, onshore and offshore. Brazil."],
#> BBOX[-80,-48,0,-42]],
#> ID["EPSG",32723]]
# import raster from r to grass
rgrass::write_RAST(x = r, flags = c("o", "overwrite", "quiet"), vname = "r")
#> SpatRaster read into GRASS using r.in.gdal from memory
# distance
lsmetrics::lsm_distance_enn(input = "r")
#> Converting zero as null
#> Identifying fragments
#> Calculating distance
#> [1] "1 of 1"
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
#> Changing the raster color
#> Cleaning data
# files
rgrass::execGRASS(cmd = "g.list", type = "raster")
#> r
#> r_distance_enn
# import from grass to r
r_distance_enn <- terra::rast(rgrass::read_RAST("r_distance_enn", flags = "quiet", return_format = "SGDF"))
#> Creating BIL support files...
#> Exporting raster as integer values (bytes=4)
#> 0% 6% 12% 18% 25% 31% 37% 43% 50% 56% 62% 68% 75% 81% 87% 93% 100%
# plot
plot(r_distance_enn, legend = FALSE, axes = FALSE, main = "Euclidean nearest neighbor distance (m)")
plot(as.polygons(r, dissolve = FALSE), lwd = .1, add = TRUE)
plot(as.polygons(r), add = TRUE)
text(r_distance_enn, cex = .5)
 # delete grassdb
unlink("grassdb", recursive = TRUE)
# delete grassdb
unlink("grassdb", recursive = TRUE)